Dealing with Legacy Apps: Remodeling the Hepatitis C Assessment
📅 02 June, 2019
•⏱️11 min read
The Hepatitis C Consultation Note was one of the first e-forms to be produced by NHS Wales Informatics Service. This digital form provides a means of capturing the clinical assessment from an outpatient appointment. It relies on data derived from two principle data sources: the being Welsh Results Reporting Service (WRRS) for pathology data as well as natively committed data via the form itself. This application is currently used in outpatient settings in NHS Wales.
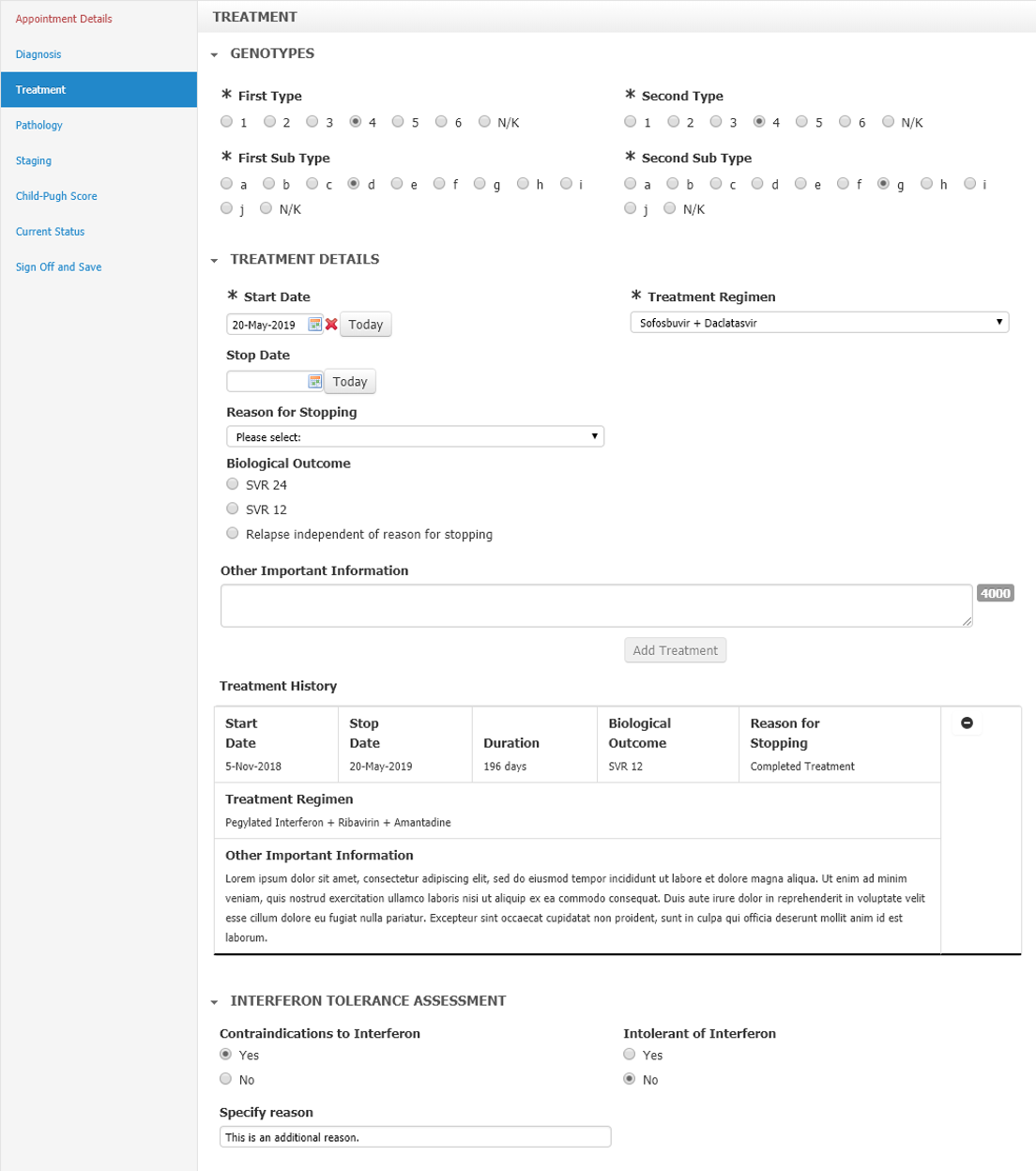 Fig 1: Example section within the Hepatitis C form.
Fig 1: Example section within the Hepatitis C form.
The goal of this exercise was to see how easily the current form could be modelled in OpenEHR. This achieves two purposes;
- tests the efficacy of the OpenEHR modelling tools, and
- assess whether the fabled idea that “the modelling has already been done” exists for this use case.
At this point, the Hepatitis C form is a legacy application i.e. it was not architected to natively store data in a structured format. That is not a major issue as the data is stored as XML and extracted from the resulting composition later. But this is a good example of many applications that are currently in use where we may want to re-engineer in future.
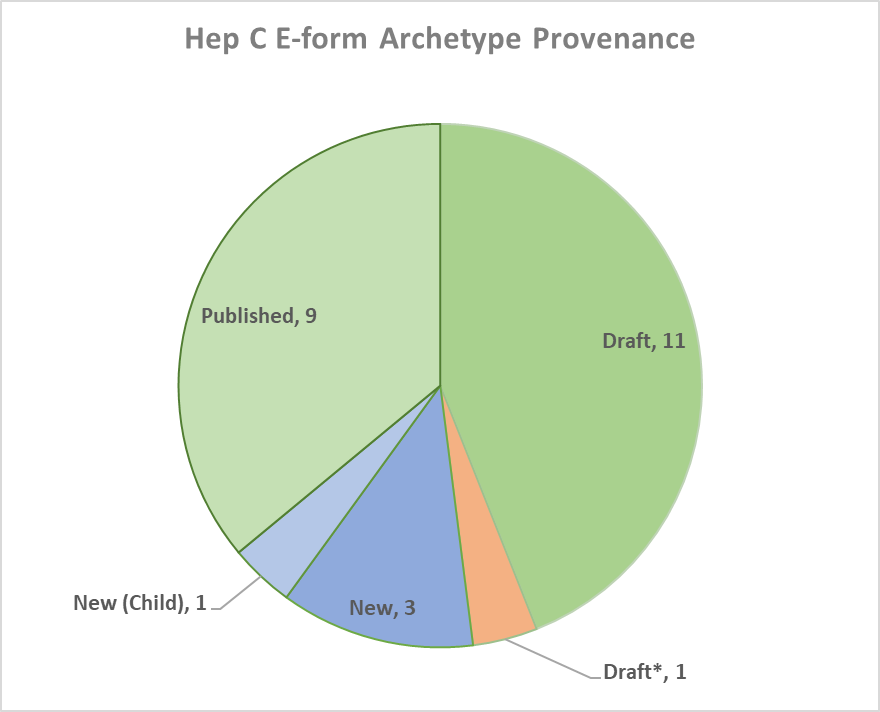 Fig 2: Devleopment status of archtypes used in the resulting OpenEHR template
Fig 2: Devleopment status of archtypes used in the resulting OpenEHR template
Initial analysis of the Hepatitis C form revealed a significant amount of reusability of previously published archetypes with 84% of the resulting template already available in the international CKM. The model set consisted of 3 new archetypes, 1 child specialisation for a previously available archetype and another draft archetype that needed to be re-modelled. 9 of the archetypes used had been published while a further 11 were in draft state.
While the general process of selecting archetypes is worthy of mention, I would like to focus upon the issues affecting two aspects of the model in particular; the specialisation of a child archetypes and where differing modelling approaches need to be considered.
Specialising the Medication Summary Archetype
The Medication Summary for Hepatitis C was developed as a child archetype. A child archetype is defined as one that wholly conforms to its parent but features non-breaking additions to the model that do not contravene its initial purpose. It is a specialism of the parent archetype that does something "extra".
The Medication Summary, as original conceived is designed to provide summary information about the administration or consumption history for a specified medication or class of medication over the individual's lifetime. It is a draft archetype published in the international CKM.
To fit the Hepatitis C form, it required the addition of “Treatment outcome” to the archetype to record a narrative description of outcome for this episode of treatment. This addition can be viewed as the lowest element within the element cluster.
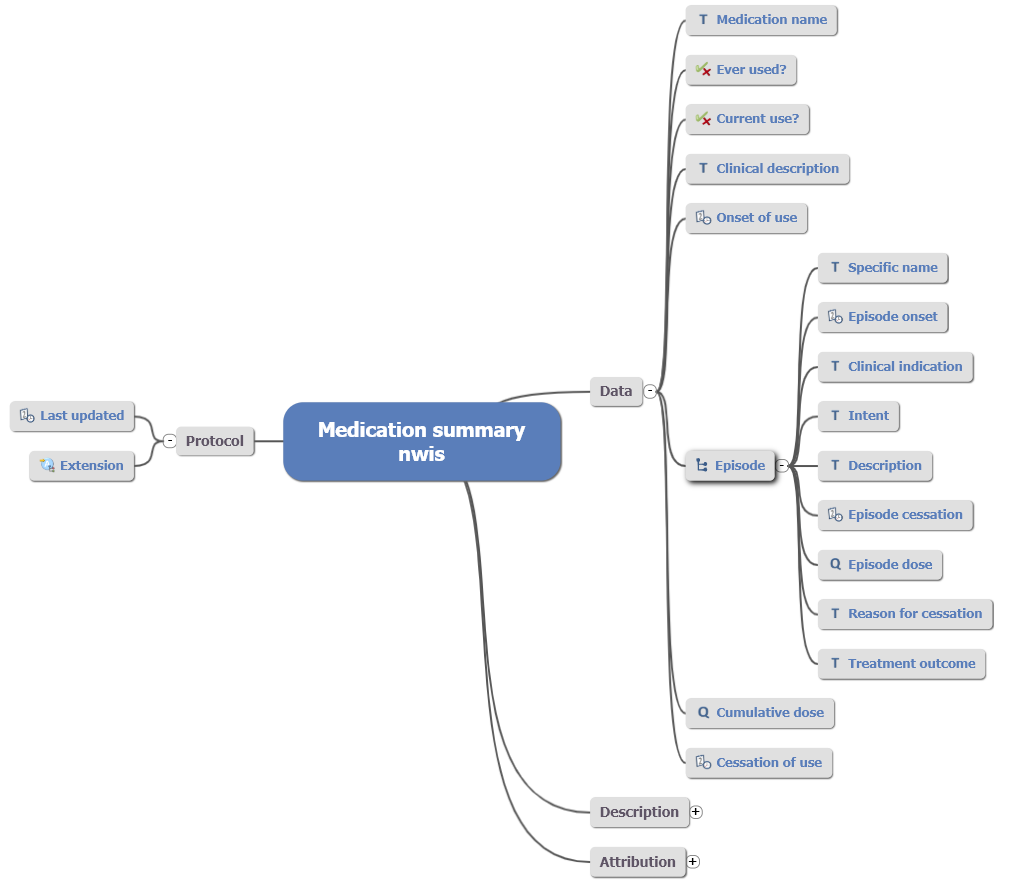 Fig 3: Child archetype adapted from orignial draft Medication Summary.
Fig 3: Child archetype adapted from orignial draft Medication Summary.
In terms of local specialisation, all future changes to the parent will cascade down to the child archetype. Additionally, this specialisation may be adopted into the parent under a review round as the provenance of this change lies within a demonstrable clinical requirement. Wider to this, the data recorded in either archetype is semantically coherent when queried (barring the specialist child element).
Archetype Specialisation Implications
When a child archetype is published, it presents the option of incorporating the new specialist elements into the parent via a change request. This represents the gradual evolution of that model as part of the review and curation process in the Clinical Knowledge Manager. The outcome of this evaluation may of course result in the change being rejected, in which case the option of maintaining the child version is available. In terms of clinical data standards, an organisation is then faced with the decision to support either or both archetypes.
If the specialist archetype is maintained for all use cases, it leads to consistency across the clinical model domain. This may be on a hospital, regional or national basis depending on what the use case is. The risk to semantic coherence is mitigated due to the nature of the child/parent archetype but this does not rule out the use of the parent solely where new use cases present.
There is of course a responsibility to ensure the maintenance of the child archetype is carried out. In an example where changes are made to the parent, there is no automatic process to subsume these within the child. The child archetype in effect contains an embedded version of the parent, and reapplying upstream changes is something that is done manually. This presents two options;
- Incorporate the changes by directly editing the archetype via the Archetype Definition Language (ADL)1.
- Create a new specialised child archetype from the updated parent i.e. repeat the original changes
Additionally, the decision to update the child archetype resides with the owner. The potential for changes that break downstream applications needs to be carefully considered although this is nothing specific to OpenEHR – it applies to all application developments where the underlying data structures change.
However, caution is recommended where specialisation occurs to ensure that a more appropriate modelling route is not ignored. An example could present itself as multiple additions which may lead to conflict with elements of the parent (e.g. use or misuse, or that parent elements require change). As a rule of thumb, a large volume of specialisations in the child archetype could mean that the wrong parent is being used.
The Child-Pugh Score: A Modelling Conflict
The Child-Pugh score is used for chronic liver disease/cirrhosis prognosis. Higher Child-Pugh scores indicate worsening liver function and give the medical and surgical teams an idea of "liver comorbidity" and "liver reserve". This can be useful when planning interventions on the liver since liver failure is the main cause of death after liver resection.
Clinical scores and measures will invariably comprise of more than one individual data points. A simple example is the Body Mass Index which requires height and weight. However, the use case described by the Child-Pugh Score created a modelling pattern conflict with two viable approaches; use an OBSERVATION based archetype or a medication CLUSTER.
Archetype Class Theory
The two types of archetype class considered for this model are an OBSERVATION and a CLUSTER.
OBSERVATIONarchetypes contain raw information. They can be interpreted as the evidence to clinical care concerning symptoms or signs, clinical findings or test results. Importantly,OBSERVATIONclass archetypes are used to record the output of direct clinical contact with the patient of subject. Simple observations include height, weight, blood pressure or other blood tests.CLUSTERarchetypes are used withinENTRYor otherCLUSTERarchetype nodes. They can be used to establish a common domain pattern that may be required in many common archetypes or clinical scenarios. They can be best described as the plug that fits an information model socket (orSLOT).
Examples include Address or Anatomical location which are generic and standardised data fragments for use in multiple scenarios. An OBSERVATION archetype for example could symptom contain the SYMPTOM/SIGN archetype. We would then know that every time this is used, data would follow a generic pattern for all symptoms and reported signs. The 'Specific details' SLOT can be used to extend the archetype to include additional, specific data elements for more complex symptoms or signs.
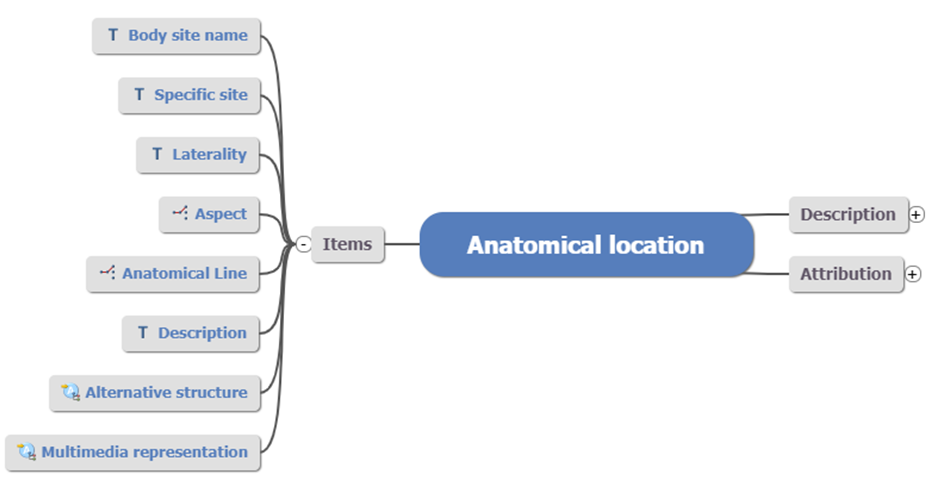 Fig 4: Anatomical location
Fig 4: Anatomical location CLUSTER archetype
In the above example, you can see generic data elements such as Laterality and Description. Using this structure over and over where appropriate provides an additional dimension to data standardisation; not just the what but how that data is to be captured and stored.
CLUSTER archetypes do not maintain the provenance of the data themselves. They have no event or timing attributes as these are derived from the ENTRY archetype that contains the given cluster (i.e. the data socket).
The below image (Fig 5) depicts the insertion of the Language cluster archetype into an Additional language slot in a template using the Marand Archetype Designer. This slot has been constrained to only accept this specific archetype which has been modelled to provide a generic and standardised data fragment for multiple use. This approach demonstrates both flexibility as well as a means of standardising patterns of clinical data. Rather than allowing any CLUSTER to fit with your model and relying instead on the knowledge of the person building the template, this allows the author to prevent potential issues in semantic coherence earlier.
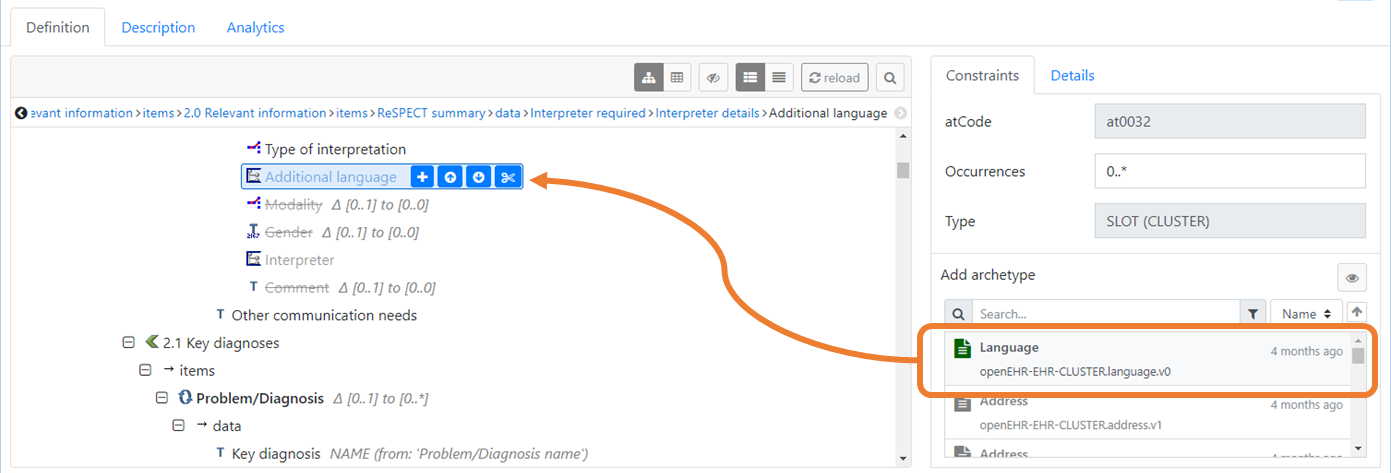 Fig 5: Example of how a
Fig 5: Example of how a CLUSTER archetype can be constrained to only "fit" a specific SLOT.
Remodelling an Archetype
A draft archetype of Child-Pugh Score was available but had not been published. This was based on the CLUSTER model. Its purpose was “to be used in the Test findings slot in a laboratory test result observation archetype or in the Specific details slot of a problem/diagnosis evaluation archetype”.
It is acceptable for a simple scale to be a cluster but anything that has a total or some interpretative finding should be an observation to ensure semantic coherence. That way, the additional attributes found as part of the reference model are maintained such as the history of the measurements. This is important and one of the key differences that marks the OpenEHR Reference Model as an essential component to the machinery that underpins a digital health record.
As a result, the Child-Pugh score was recreated as an OBSERVATION archetype:
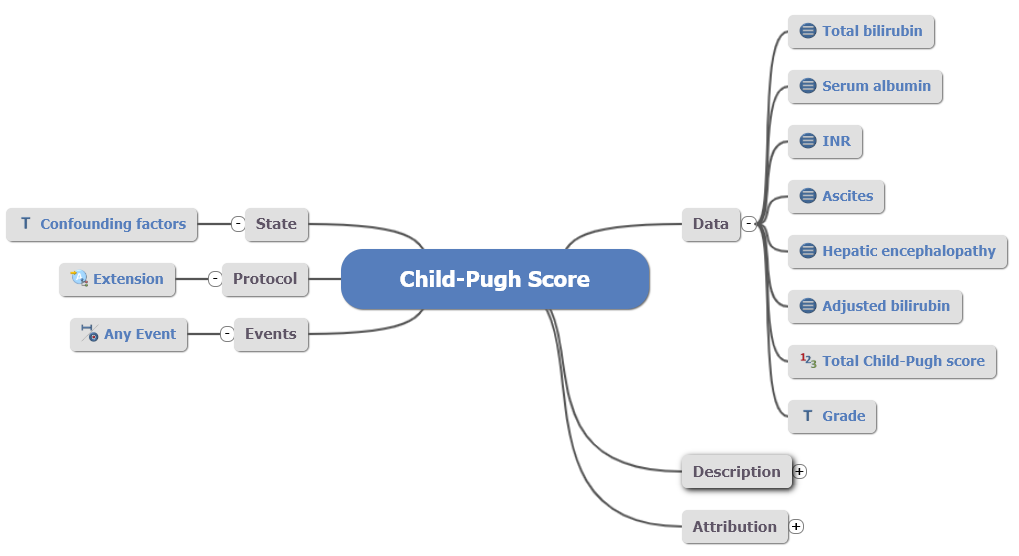 Fig 6: Remodelled Child-Pugh Score
Fig 6: Remodelled Child-Pugh Score OBSERVATION archetype.
In the case of a previously modelled archetype, the EHRScape Archetype Designer tooling offers a flexible capability to copy and paste elements between archetypes. This meant that the revised OBSERVATION based archetype for Child-Pugh Score could be recreated within minutes, maintaining much of the previous work that had been done. This has important implication for clinical modelling workflow where common patterns are found and need to be rapidly replicated.
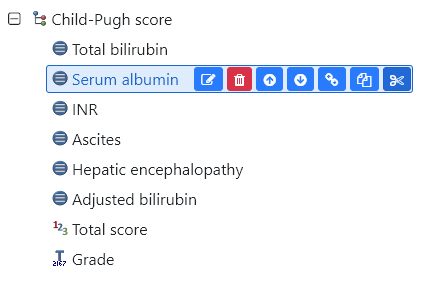 Fig 7: Wow. Cut and paste an archetype element? #TIMESAVER
Fig 7: Wow. Cut and paste an archetype element? #TIMESAVER
Modelling took around 2 days to complete and resulted in a template which is now available on the Apperta CKM for review. In addition to this, the EHRScape form designer was able to be tested to replicate the current e-form. It is clear from the current tooling that this functionality is intended for simple forms albeit with potentially complex logic but I am lead to believe a revised form designer is imminent.
Using Clinical Terminology
For the Hepatitis C form, the mapping to Snomed CT proved to be patchy. Where the clinical requirement states a range of genotype subtypes (e.g. a, b, c through to ‘Not Known’ - see Fig 1, above), the combination of the Type and Subtype are also individual codes in the Snomed hierarchy (e.g. Hepatitis C virus subtype 2c is code 603419009).
However not all of the combinations needed in the form can be found in Snomed CT which presents an ontological gap. Therefore a combination of Snomed CT codes for the type were used (e.g. 603426009 | Hepatitis C virus genotype 5 (organism)) and a local code list for subtype (e.g. a, b, c etc). This is an all too common example in my experience and proves that Snomed CT still needs active and ongoing work to support the ambition of ubiquitous structured and coded data in the digital record.
Closing Thoughts
 From the outset I expected that there would be a good degree of reusability, however I was presently surprised at the extent of how many archetypes were ready to go. That did not mean that the modelling process was perfect or easy but overall it was a valuable learning exercise and produced a very usable template.
From the outset I expected that there would be a good degree of reusability, however I was presently surprised at the extent of how many archetypes were ready to go. That did not mean that the modelling process was perfect or easy but overall it was a valuable learning exercise and produced a very usable template.
Unearthing archetypes from non-CKM sources was also interesting but proved my hunch that a lot of work has gone into models that may be consistently "work in progress". It helps that I know people… But I believe the OpenEHR community faces a challenge where draft archetypes, and a multitude of “local” use cases still need to be visible and open for curation. This is not an easy endeavour and takes a lot of effort especially where the local requirements are very specific (be that country/area specific or for niche clinical domains). It is an evolutionary process and one that will gather pace with a greater number of models becoming published. The evidence presented here of a seemingly niche NHS Wales application example is testament to the growing knowledge base present in both the UK based Apperta CKM and the international one. It is both feasible and realistic to use OpenEHR as a modelling base.
Finally, I am hoping that the specialisation I have suggested for Medication Summary can be incorporated into the international archetype, and further to this that the percentage of draft to published archetypes can also be increased over time. As a result, I will be adopting these archetypes to support the ongoing curation process. But the important point is that most of the models already exist which makes the task of standardising clinical data much easier.
Photo credit: Ben White
-
This is made easier with ADL2 although I have yet to dig around into the specifics. It’s on my To Do list!
↩